Research |
NGS facility |
Publications |
Laboratory |
People |
Photos |
Science |
High-throughput "next-generation" sequencing capability at Wright State |
Supported by a recent NSF Major Research Instrumentation Grant awarded to Dr. Oleg Paliy and Wright State University, we have established a high-throughput sequencing capability based on the Ion Torrent next-generation sequencer. Ion Torrent Personal Genome Machine (PGM) Sequencer is a third-generation sequencer that is based on the use of a massively parallel array of proprietary semiconductor sensors to perform direct real-time measurements of the hydrogen ions released during DNA replication. The unique combination of fluidics, micromachining, and semiconductor technology enables the direct translation of genetic information (DNA) to digital information (DNA sequence), rapidly generating large quantities of high quality data at low costs. The NGS equipment is housed in the Wright State Center for Genomics Research and comprises Ion Torrent Personal Genome Machine (PGM) Sequencer and Server, Ion Torrent One Touch sample preparation system, Pippin Prep DNA fragment selection system, and Qubit fluorometer. We are happy to extend high-throughput sequencing capabilities to other researchers on campus and from outside organizations and institutions. Please contact Dr. Oleg Paliy ( |
|
|||||||||||||||||
High-throughput sequencing has revolutionized the current research approaches in many fields of Biological Sciences. Because of the ability of the next generation sequencing (NGS) technologies to output millions of nucleotide sequence reads in a single run, NGS methods are dramatically advancing our ability to comprehensively interrogate the nucleic acid-based information in cell(s) at unparalleled resolution and depth. The underlying sequencing technology of Ion Torrent PGM exploits a well-characterized biochemical process: when a nucleotide is incorporated into a strand of DNA by a polymerase, a hydrogen ion (H+) is released as a byproduct. This hydrogen ion carries a charge which the PGM ion sensor can detect as a change in the pH in the well. A high-density array of wells on the semiconductor chip provides millions of individual reactors while integrated fluidics allow reagents to flow over the sensor array. As the sequencer floods the chip with one nucleotide after another, any nucleotide added to a DNA template will be detected as a voltage change, and the PGM will call the base. If a nucleotide is not a match for a particular template, no voltage change will be detected and no base will be called for that template. |
|||||||||||||||||
A range of experimental approaches that are enabled with the use of Ion Torrent PGM include:
Choice of sequencing chips |

|
||||||||||||||||
|
Currently, three chips are available for the use on Ion Torrent PGM. This provides a choice of the most appropriate chip to use in a project based on the number of reads and/or sequence output required. |
|||||||||||||||||
|

|
||||||||||||||||
Multiplexing Multiplexing allows multiple samples to be processed simultaneously on the same chip, which reduces costs, time, and labor. During library preparation, specific primers that contain a short barcode sample identification sequence are attached to DNA fragments. The individual samples are then mixed together and this multiplexed sample is processed further. During data analysis, barcodes are read and used as indexing tags to identify which sample each read belongs to. Workflow 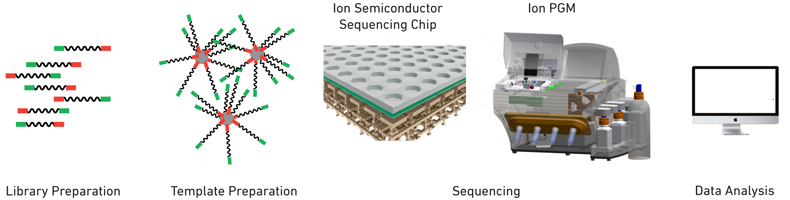 |
|||||||||||||||||
|
|
All services are customizable depending on the specifics of the user project, required depth of coverage, and number of samples to be processed. Below we provide estimated ranges for each type of service and analysis:
|
|
|
|
|
 Ion Torrent PGM Sequencer is the core component of the NGS system. PGM Sequencer is a benchtop semiconductor-based platform that performs a sequencing run in two to five hours (depends on read length). It has a small benchtop footprint and weight, uses low cost sequencing reagents, and is semi-automated in use. The Sequencer utilizes microfluidics chips comprising millions of individual wells where template-containing Ion Spheres are added. The sequence read length can be varied by the use of different sample preparation and sequencing kits from 100 to 200 to 400 nct. |
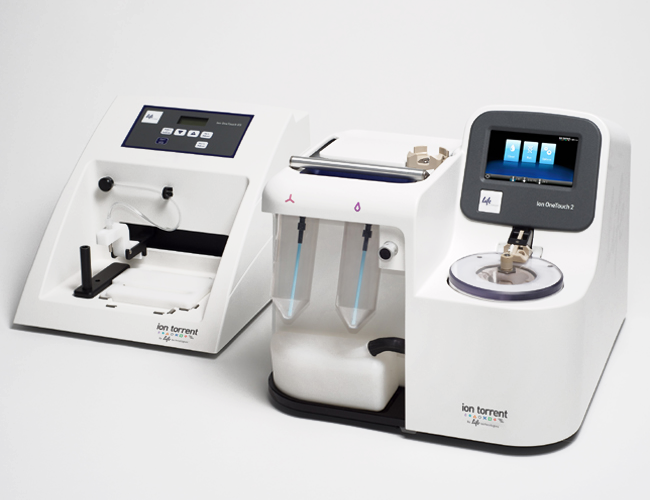 This instrument enables reproducible template preparation for the PGM Sequencer in about 4-5 hours. It is comprised of two components: (i) Ion One Touch Instrument, which utilizes in-line PCR, combining amplification and templated Ion Sphere recovery into a single automated process; (ii) Ion One Touch Enrichment System, which provides an automated and highly reproducible magnetic particle-based enrichment procedure for isolating template-positive Ion Sphere particles. The system supports all 3 chips that are currently available as well as multi-template library construction. |
|
|
 This device is a small tabletop instrument that automates the size selection and collection of DNA fragments for PGM Sequencer libraries. DNA samples are resolved electrophoretically on the pre-cast agarose gels, and the fragments of correct size are eluted into a special chamber where they can later be collected with a pipet tip. This allows the elution of a user-defined range of DNA fragment sizes, which produces uniform amplicon size pool and significantly reduces contamination with small fragments. |
 The Qubit 2.0 Fluorometer quantitates DNA, RNA, and protein with high accuracy and sensitivity. It utilizes specifically designed fluorometric Molecular Probes dyes to quantitate biomolecules of interest. These fluorescent dyes emit signals only when bound to specific target molecules (dsDNA, ssDNA, RNA, etc), even at low concentrations. For example, Qubit allows a precise quantitation of dsDNA after PCR reaction and the readings are not influenced by nucleotides and other contaminants. |
 Available capabilities and types of analyses
Available capabilities and types of analyses